Reproducible Experiments in Machine Learning
How to make scientific results reproducible using containers
 Photo by chuttersnap on Unsplash
Photo by chuttersnap on UnsplashReplication Crisis
Today not only the economy but also science is working in a breakneck pace. Even more accelerated through the current pandemic, the iteration time of new scientific research is short and not much time for peer review is available.
Good practice in science (and life in general) is the replication of results: to check for correctness or to facilitate understanding. This is crucial in a peer review process, having an ever increasing amount of scientific papers with questionable quality. One big problem is therefore the lack of replicable results also known as the replication crisis. The term covers many facets of this problem in different scientific disciplines. Specifically in machine learning many results and comparisons are questionable. A recent study tried to replicate results in scientific papers and was only successful on average 40% of the time.
Current problems
There are many frustrating aspects I encountered when reading papers in machine learning:
1. No public data used
While I understand the reasons for it a study should not solely rely on private data to highlight its merits.
2. No source code
While many scientists are not software engineers and are shy about sharing their paper-deadline scripts it should at least be part of the requirements in journals to share the code. It is not reasonable for other peers to replicate results by implementing algorithms themselves.
3. High install/usage barriers
If the source code is available, necessary declarations of dependencies can be missing which requires installing those manually (if possible). This problem gets worse with age of the publication, as newer versions of programming languages or libraries are not always backwards compatible.
4. No replicable experimental setup
Even if 2. and 3. are fulfilled, sometimes the experimental scripts are missing. While most parameters should be part of the scientific manuscript itself, sometimes authors forget to mention crucial preprocessing steps or parameters. If the complete experimental script is available, which was used to produce the results in the paper, this problem would be impossible.
(5. Lack of necessary resources to replicate models)
Even if all the former things are provided, another problem, specifically in deep learning, is the necessary amount of resources to train a model. Many big players (Google et al.) in this area have nearly unlimited GPU resources available which is unattainable for many research institutions. This also often leads to the question, if stated improvements are based on architectural changes or on more time for training.
Towards reproducible science and experiments
While aspects 1. and 2. are getting better in my opinion a solution to 5. is still not clear to me. On the other hand 3. and 4. can be improved but are still lacking in academia as they require skills in software engineering and development most often found in industry applications.
In the following I will describe how I made the experiments in my newest scientific preprint reproducible.
The source code of the algorithm is available on GitHub.
In my case I am using Python for the implementation. There exist (too) many approaches for Python to declare dependencies. A new alternative which tries to encapsulate the best ideas of all before is the tool Poetry, which allows the declaration of general requirements and also specific versions.
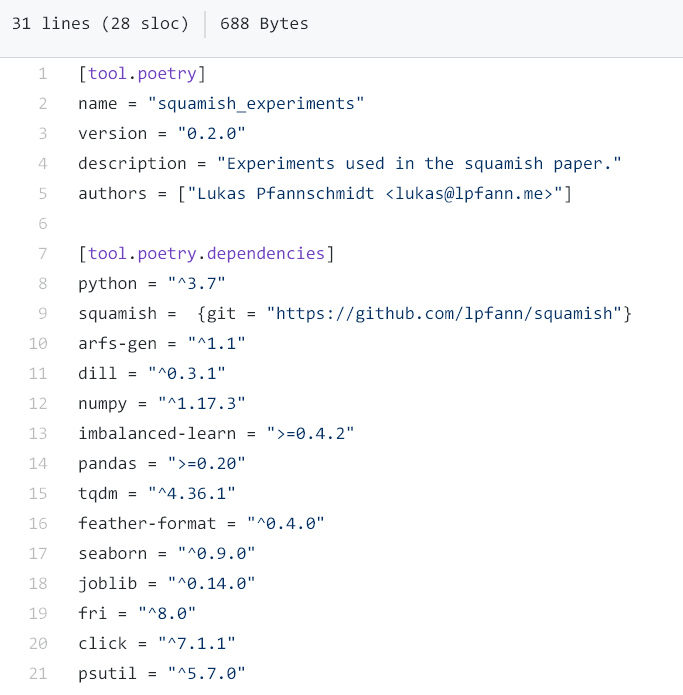
In addition, Poetry supports the automatic creation of virtual environments which encapsulate these specific dependencies, even if the global Python environment is widely different to the original creators. These environments are defined by hashes which Poetry automatically derives and are located in the poetry.lock file.
To get another layer of encapsulation we also utilize Containers made popular under the name Docker. While Poetry encapsulates Python environments, containers can encapsulate the complete operating system. This makes it possible to run experiments even many years in the future with the same global software stack.
We provide a GitHub repository for all experiments with a Dockerfile included, which is basically a recipe list for all software needed (including the OS).
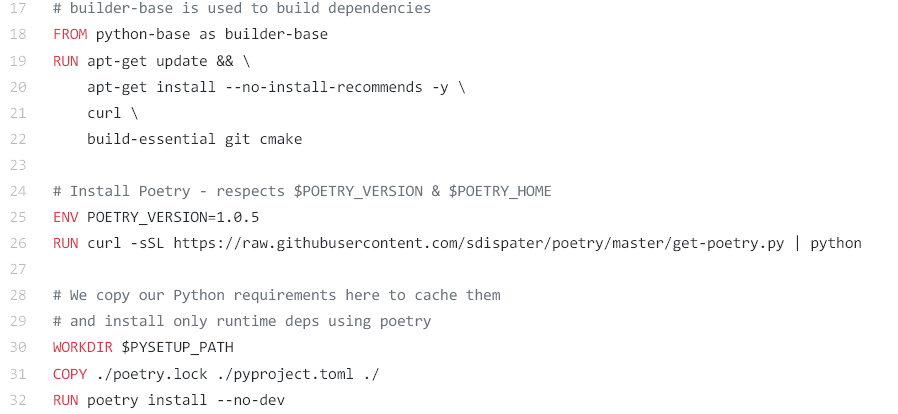
In short the Dockerfile instructs the container builder to use a specific OS and Poetry to install all Python dependencies and create an image. One can also execute these instructions beforehand to create a container image and upload it to a public repository, which makes building unnecessary.
Now we can provide the potential reviewer or user with the following instructions which automatically perform replication:
Instructions for replication
To replicate the experimental results of the paper (figure and tables)
1. Get container image
Build the image yourself with
docker build -t squamish_experiments .
or pull it from DockerHub
docker pull mirek1337/squamish_experiments
2. Run container
docker run -v ./tmp:/exp/tmp:Z -v ./output:/exp/output:Z -it squamish_experiments make
which calls make inside the container to execute all experiments in the Makefile.
After the experiments are done (can take several hours) the output should end up in the ./output folder.
It’s also possible to change the following parameters as environment variables in the docker command via the -e option:
Defaults used in paper
SEED= 123REPEATS= 10N_THREADS= 1
Conclusion
While working with containers is still new for many scientists, the advantages are big. One can not expect everybody to know the tools in detail, and more work in usable abstractions is needed. For now there are project templates or libraries available with which make this work easier.